It’s nearly been two years since I defended my PhD thesis! On top of allowing me to call myself doctor, having a PhD in statistics gives me the honour to feature in the data of the Mathematics Genealogy Project. Today, I decided to webscrape my mathematical ancestors.
My blogging ancestors?
Before blogging, following my own advice I googled my idea and found these similar efforts:
-
Nathalie Vialaneix scraped her own mathematical tree using the R
XMLpackage and drew it using theigraphpackage andGraphViz. My post is basically a remix of her post. -
Heike Hofmann started writing a package to scrape the Mathematical Genealogy Project, and apparently abandoned it.
-
Thibault Vatter published a GitHub repo mixing R, Python and a tool called Scrapy to scrape the website.
I wrote my own version of the webscraping because I wanted to use rvest like I usually do these days.
Scraping the Mathematical Genealogy Project
Now that I have added tags to all my old blog posts, I can look back at my previous webscraping efforts and use my recent scripts… as well as see how much progress I made since my reckless webscraping days, when I didn’t check I was allowed to webscrape, and when I used on string manipulation rather than XPath and friends.
Nowadays before webscraping I use the rOpenSci robotstxt package to read what the website owners will let me do.
> robotstxt::robotstxt("https://www.genealogy.math.ndsu.nodak.edu")$text
User-agent: msnbot
Crawl-delay: 30
User-agent: Browsershots
Disallow:
User-agent: *
Disallow: /submit-data.php
> robotstxt::paths_allowed("https://www.genealogy.math.ndsu.nodak.edu/id.php")
www.genealogy.math.ndsu.nodak.edu
[1] TRUE
From the above I deduced that
-
there is no recommended delay for scrapers that are not msnbot, so I used 5 seconds between calls, which seemed lagom.
-
I was allowed to scrape mathematicians’ pages. Hooray!
Then, I looked at the code of my most recent webscraping blog post that was building on code by Bob Rudis: after looking at the source of one mathematician’s page, I used html_attrs and html_text to extract the elements I needed from each page.
.get_advisors <- function(id_string, sleep_time){
# small break to be nice
Sys.sleep(sleep_time)
# try to get the page
page <- glue::glue("https://www.genealogy.math.ndsu.nodak.edu/{id_string}") %>%
httr::GET()
# try until it works but not more than 5 times
try <- 1
while(httr::status_code(page) != 200 & try <= 5){
Sys.sleep(sleep_time)
page <- glue::glue("https://www.genealogy.math.ndsu.nodak.edu/{id_string}") %>%
httr::GET()
try = try + 1
}
# Now get student's name
student_name <- httr::content(page) %>%
rvest::xml_nodes(xpath = '//h2[@style="text-align: center; margin-bottom: 0.5ex; margin-top: 1ex"]') %>%
rvest::html_text() %>%
stringr::str_remove("\\\n")
# Get all nodes corresponding to advisors
# Thanks to their... formatting but it works
all_advisors <- httr::content(page) %>%
rvest::xml_nodes(xpath = "//p[@style='text-align: center; line-height: 2.75ex']") %>%
rvest::html_nodes("a")
# Export results
tibble::tibble(student_name = student_name,
id_string_student = id_string,
name = purrr::map_chr(all_advisors, rvest::html_text),
id_string = purrr::map_chr(all_advisors, rvest::html_attr,
"href"))
}
Now, since sometimes advisors will be encountered more than once in the data, I used memoise to create a handy function wrapper that will cache results.
# Cache results in case a mathematician comes up several times
get_advisors <- memoise::memoise(.get_advisors)
Finally, to get all my ancestors, I had to iteratively get the ancestors of each of my ancestors… until when? Heike Hofmann wrote a function working a given number of steps, Nathalie Vialaneix stopped when there was no advisor on an advisor’s page, I stopped when the data.frame stopped growing because that’s what I found the easiest to implement.
me <- "id.php?id=207686"
# initial data.frame
df <- get_advisors(me, 5)
new_df <- df
keep_growing <- TRUE
while(keep_growing){
# get size to compare to size after a bit more scraping
nrow1 <- nrow(df)
# get advisors for all new lines
# from the previous iterations
new_df <- purrr::map_df(new_df$id_string, get_advisors, sleep_time = 30)
df <- unique(rbind(df, new_df))
# if the data.frame didn't grow, stop
if(nrow(df) == nrow1){
keep_growing <- FALSE
}
}
# save results
readr::write_csv(df, "math_ancestry.csv")
I tested the above on very ancient mathematicans who didn’t have too many ancestors to check it was working, and stopping.
At the end of my data gathering, I had a nice table of 261 mathematicians including yours truly! By the way, the Mathematical Genealogy Project maintainers state “Throughout this project when we use the word “mathematics” or “mathematician” we mean that word in a very inclusive sense. Thus, all relevant data from mathematics education, statistics, computer science, or operations research is welcome.” which is the reason why I feel fine calling myself a mathematician in this post.
Showing (off) my mathematical family tree
The approach I used below, defining nodes and edges for integration by DiagrammeR before exporting to igraph and then to GraphViz (not an R package, but interfaced by both DiagrammeR and igraph), might seem a bit complicated since the DiagrammeR package itself exports to GraphViz format… but not with the classic default look I liked on Nathalie’s Vialaneix blog.
library("magrittr")
library("DiagrammeR")
# create nodes
labels <- unique(c(df$student_name, df$name))
nodes_df <- create_node_df(n = length(labels))
nodes_df$label <- labels
# create edges
edges_df <- df[, c("name", "student_name")]
edges_df <- dplyr::left_join(edges_df, nodes_df,
by = c("name" = "label"))
edges_df <- dplyr::rename(edges_df, from = id)
edges_df <- dplyr::left_join(edges_df, nodes_df,
by = c("student_name" = "label"))
edges_df <- dplyr::rename(edges_df, to = id)
# special character that'd make GraphViz throw an error
nodes_df <- dplyr::mutate(nodes_df,
label = stringr::str_replace_all(label, "'", " "))
# create a DiagrammeR dgr_graph object
dgr <- create_graph(nodes_df = nodes_df,
edges_df = edges_df[, c("to", "from")],
directed = TRUE)
# export the object to igraph format
# and then write it to a GraphViz DOT file
DiagrammeR::to_igraph(dgr) %>%
igraph::write.graph(file = "graph.dot",
format = "dot")
I vaguely got the ambition to use some sort of htmlwidget to have a zoomable and pretty network but didn’t want to spent too much time doing it, so PNGs it is! Here is how I hacked my way to a PNG.
DiagrammeR::grViz("graph.dot") %>%
htmlwidgets::saveWidget("lala.html")
webshot::webshot("lala.html",
selector="#htmlwidget_container",
file = "tree.png",
zoom = 10)
file.remove("lala.html")
I then cropped two zooms by hand!
So here is my whole tree…

It is unreadable unless you open it in its own tab and zoom. Like Nathalie Vialaneix, I think I only have male mathematical ancestors.
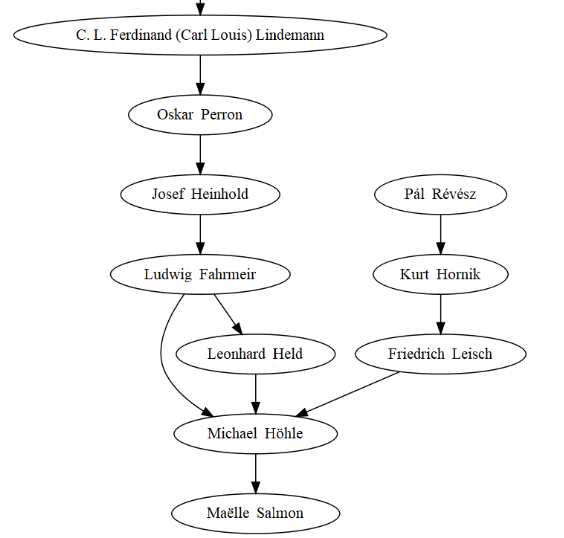
The most recent part of the tree obviously features my PhD advisor Michael Höhle, who by the way has a very smart statistics blog! I also see I’m related to CRAN’s Kurt Hornik, which doesn’t mean I get my packages on CRAN whilst bypassing gatekeeping though.
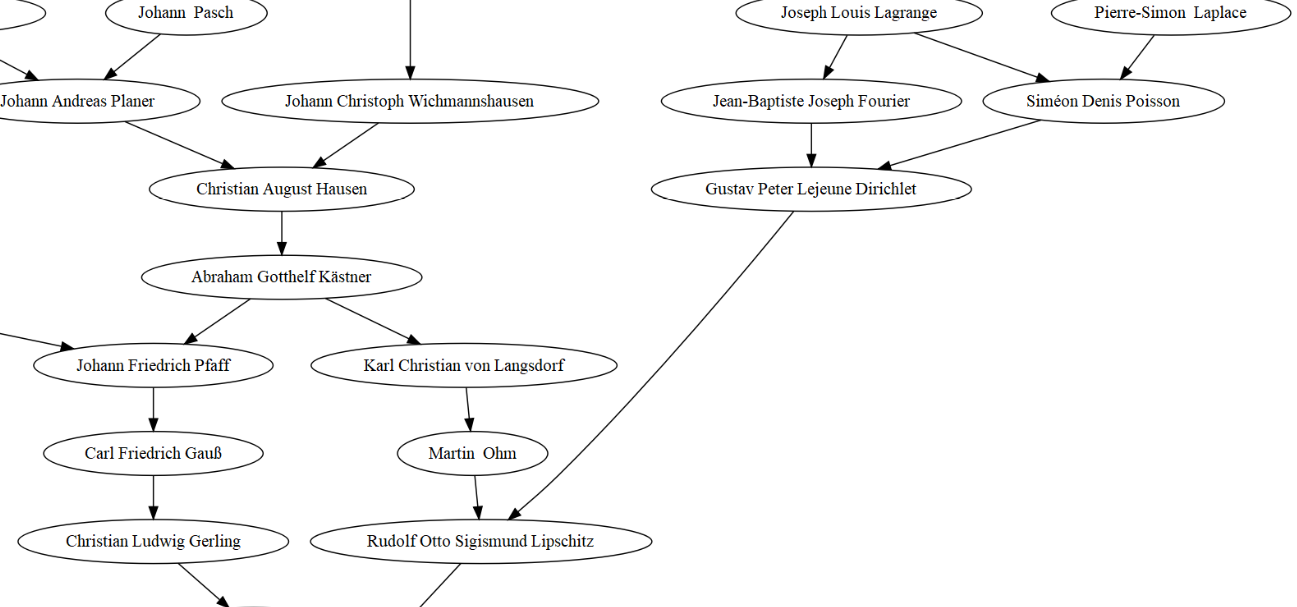
I was then very pleased to recognize some famous mathematicians’ names, not the most ancient ones I’m afraid, but look, Poisson! I was very excited about this because my PhD thesis featured count data, and because Poisson means fish in French… So maybe I’m a fish after all. Besides, seeing Carl Friedrich Gauß also made me happy… I had no idea the picture below was actually a family portrait!
Growing this post?
In conclusion, I scraped and drew my mathematical family tree using data from the Mathematical Genealogy Project. Extensions of my post could include making a package like Heike Hofmann’s one to make it easier for anyone to get their data; preparing a nicer visualization (note that you can buy posters to support the project as explained on this page), and getting more data from each mathematician’s page to draw once’s ancestry on a map, summarize thesis topics with tidy text analysis… Now, I’ll probably remain the black sheep of my mathematical family by blogging about kitsch plots!