I’ll admit I didn’t really know who Bill Nye was before yesterday. His name sounds a bit like Bill Nighy’s, that’s all I knew. But well science is all around and quite often scientists on Twitter start interesting campaigns. Remember the #actuallylivingscientists whose animals I dedicated a blog post? This time, the Twitter campaign is the #BillMeetScienceTwitter hashtag with which scientists introduce themselves to the famous science TV host Bill Nye. Here is a nice article about the movement.
Since I like surfing on Twitter trends, I decided to download a few of these tweets and to use my own R interface to the Monkeylearn machine learning API, monkeylearn (part of the rOpenSci project!), to classify the tweets in the hope of finding the most represented science fields. So, which science is all around?
Getting the tweets
It might sound a bit like trolling by now, but if you wanna get Twitter data, I recommend using rtweet because it’s a good package and because it’s going to replace twitteR which you might know from other blogs.
I only keep tweets in English, and moreover original ones, i.e. not retweets.
library("rtweet")
billmeet <- search_tweets(q = "#BillMeetScienceTwitter", n = 18000, type = "recent")
billmeet <- unique(billmeet)
billmeet <- dplyr::filter(billmeet, lang == "en")
billmeet <- dplyr::filter(billmeet, is_retweet == FALSE)
I’ve ended up with 2491 tweets.
Classifying the tweets
I’ve chosen to use this taxonomy classifier which classifies text according to generic topics and had quite a few stars on Monkeylearn website. I don’t think it was trained on tweets, and well it wasn’t trained to classify science topics in particular, which is not optimal, but it had the merit of being readily available. I’ve still not started training my own algorithms, and anyway, if I did I’d start by creating a very crucial algorithm for determining animal fluffiness on pictures, not text mining stuff. This was a bit off topic, let’s go back to science Twitter!
When I decided to use my own package I had forgotten it took charge of cutting the request vector into groups of 20 tweets, since the API only accept 20 texts at a time. I thought I’d have to do that splitting myself, but no, since I did it once in the code of the package, I’ll never need to write that code ever again. Great feeling! Look at how easy the code is after cleaning up the tweets a bit! One just needs to wait a bit before getting all results.
output <- monkeylearn::monkeylearn_classify(request = billmeet$text,
classifier_id = "cl_5icAVzKR")
str(output)
## Classes 'tbl_df', 'tbl' and 'data.frame': 4466 obs. of 4 variables:
## $ category_id: int 64638 64640 64686 64696 64686 64687 64689 64692 64648 64600 ...
## $ probability: num 0.207 0.739 0.292 0.784 0.521 0.565 0.796 0.453 0.301 0.605 ...
## $ label : chr "Computers & Internet" "Internet" "Humanities" "Religion & Spirituality" ...
## $ text_md5 : chr "f7b28f45ea379b4ca6f34284ce0dc4b7" "f7b28f45ea379b4ca6f34284ce0dc4b7" "b95429d83df2cabb9cd701a562444f0b" "b95429d83df2cabb9cd701a562444f0b" ...
## - attr(*, "headers")=Classes 'tbl_df', 'tbl' and 'data.frame': 0 obs. of 0 variables
In the output, the package creator decided not to put the whole text corresponding to each line but its digested form itself, digested by the MD5 algorithm. So to join the output to the tweets again, I’ll have to first digest the tweet, which I do just copying the code from the package. After all I wrote it. Maybe it was the only time I successfully used vapply in my whole life.
billmeet <- dplyr::mutate(billmeet, text_md5 = vapply(X = text,
FUN = digest::digest,
FUN.VALUE = character(1),
USE.NAMES = FALSE,
algo = "md5"))
billmeet <- dplyr::select(billmeet, text, text_md5)
output <- dplyr::left_join(output, billmeet, by = "text_md5")
Looking at this small sample, some things make sense, other make less sense, either because the classification isn’t good or because the tweet looks like spam. Since my own field isn’t text analysis, I’ll consider myself happy with these results, but I’d be of course happy to read any better version of it.
As in my #first7jobs, I’ll make a very arbitrary decision and filter the labels to which a probability higher to 0.5 was attributed.
output <- dplyr::filter(output, probability > 0.5)
This covers 0.45 of the original tweets sample. I can only hope it’s a representative sample.
How many labels do I have by tweet?
dplyr::group_by(output) %>%
dplyr::summarise(nlabels = n()) %>%
dplyr::group_by(nlabels) %>%
dplyr::summarise(n_tweets = n()) %>%
knitr::kable()
| nlabels | n_tweets |
|---|---|
| 1415 | 1 |
Perfect, only one.
Looking at the results
I know I suck at finding good section titles… At least I like the title of the post, which is a reference to the song Bill Nighy, not Bill Nye, sings in Love Actually. My husband assumed that science Twitter has more biomedical stuff. Now, even if my results were to support this fact, note that this could as well be because it’s easier to classify biomedical tweets.
I’ll first show a few examples of tweets for given labels.
dplyr::filter(output, label == "Chemistry") %>%
head(n = 5) %>%
knitr::kable()
| category_id | probability | label | text_md5 | text |
|---|---|---|---|---|
| 64701 | 0.530 | Chemistry | e82fc920b07ea9d08850928218529ca9 | Hi @billnye I started off running BLAST for other ppl but now I have all the money I make them do my DNA extractions #BillMeetScienceTwitter |
| 64701 | 0.656 | Chemistry | d21ce4386512aae5458565fc2e36b686 | .@uw’s biochemistry dept - home to Nobel Laureate Eddy Fischer & ZymoGenetics co founder Earl Davie… https://t.co/0nsZW3b3xu |
| 64701 | 0.552 | Chemistry | 1d5be9d1e169dfbe2453b6cbe07a4b34 | Yo @BillNye - I’m a chemist who plays w lasers & builds |
| 64701 | 0.730 | Chemistry | 1b6a25fcb66deebf35246d7eeea34b1f | Meow @BillNye! I’m Zee and I study quantum physics and working on a Nobel prize. #BillMeetScienceTwitter https://t.co/oxAZO5Y6kI |
| 64701 | 0.873 | Chemistry | 701d8c53e3494961ee7f7146b28b9c8c | Hi @BillNye, I’m a organic chemist studying how molecules form materials like the liquid crystal shown below.… https://t.co/QNG2hSG8Fw |
dplyr::filter(output, label == "Aquatic Mammals") %>%
head(n = 5) %>%
knitr::kable()
| category_id | probability | label | text_md5 | text |
|---|---|---|---|---|
| 64609 | 0.515 | Aquatic Mammals | f070a05b09d2ccc85b4b1650139b6cd0 | Hi Bill, I am Anusuya. I am a palaeo-biologist working at the University of Cape Town. @BillNye #BillMeetScienceTwitter |
| 64609 | 0.807 | Aquatic Mammals | bb06d18a1580c28c255e14e15a176a0f | Hi @BillNye! I worked with people at APL to show that California blue whales are nearly recovered #BillMeetScienceTwitter |
| 64609 | 0.748 | Aquatic Mammals | 1ca07aad8bc1abe54836df8dd1ff1a9d | Hi @BillNye! I’m researching marine ecological indicators to improve Arctic marine monitoring and management… https://t.co/pJv8Om4IeI |
| 64609 | 0.568 | Aquatic Mammals | a140320fcf948701cfc9e7b01309ef8b | More like as opposed to vaginitis in dolphins or chimpanzees or sharks #BillMeetScienceTwitter https://t.co/gFCQIASty1 |
| 64609 | 0.520 | Aquatic Mammals | 06d1e8423a7d928ea31fd6db3c5fee05 | Hi @BillNye I study visual function in ppl born w/o largest connection between brain hemispheres #callosalagenesis… https://t.co/WSz8xsP38R |
dplyr::filter(output, label == "Internet") %>%
head(n = 5) %>%
knitr::kable()
| category_id | probability | label | text_md5 | text |
|---|---|---|---|---|
| 64640 | 0.739 | Internet | f7b28f45ea379b4ca6f34284ce0dc4b7 | @BillNye #AskBillNye @BillNye join me @AllendaleCFD. More details at https://t.co/nJPwWARSsa |
#BillMeetScienceTwitter | | 64640| 0.725|Internet |b2b7843dc9fcd9cd959c828beb72182d |@120Stat you could also use #actuallivingscientist #womeninSTEM or #BillMeetScienceTwitter to spread the word about your survey as well | | 64640| 0.542|Internet |a357e1216c5e366d7f9130c7124df316 |Thank you so much for the retweet, @BillNye! <U+263A><U+FE0F> I’m excited for our next generation of science-lovers!… https://t.co/B3iz3KVCOQ | | 64640| 0.839|Internet |61712f61e877f3873b69fed01486d073 |@ParkerMolloy Hi @BillNye, Im an elem school admin who wants 2 bring in STEM/STEAM initiatives 2 get my students EX… https://t.co/VMLO3WKVRv | | 64640| 0.924|Internet |4c7f961acfa2cdd17c9af655c2e81684 |I just filled my twitter-feed with brilliance. #BIllMeetScienceTwitter |
Based on that, and on the huge number of internet-labelled tweets, I decided to remove those.
library("ggplot2")
library("viridis")
label_counts <- output %>%
dplyr::filter(label != "Internet") %>%
dplyr::group_by(label) %>%
dplyr::summarise(n = n()) %>%
dplyr::arrange(desc(n))
label_counts <- label_counts %>%
dplyr::mutate(label = ifelse(n < 5, "others", label)) %>%
dplyr::group_by(label) %>%
dplyr::summarize(n = sum(n)) %>%
dplyr::arrange(desc(n))
label_counts <- dplyr::mutate(label_counts,
label = factor(label,
ordered = TRUE,
levels = unique(label)))
ggplot(label_counts) +
geom_bar(aes(label, n, fill = label), stat = "identity")+
scale_fill_viridis(discrete = TRUE, option = "plasma")+
theme(axis.text.x = element_text(angle = 90,
hjust = 1,
vjust = 1),
text = element_text(size=25),
legend.position = "none")
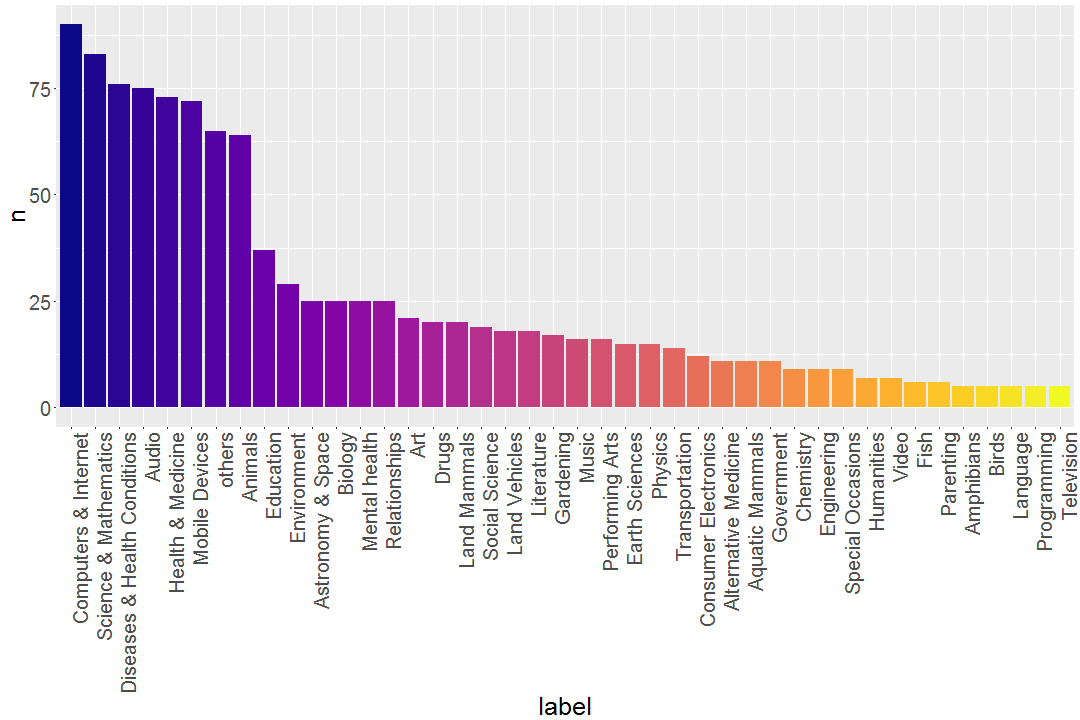
In the end, I’m always skeptical when looking at the results of such classifiers, and well at the quality of my sample to begin with – but then I doubt there ever was a hashtag that was perfectly used to only answer the question and not spam it and comment it (which is what I’m doing). I’d say it seems to support my husband’s hypothesis about biomedical stuff.
I’m pretty sure Bill Nye won’t have had the time to read all the tweets, but I think he should save them, or at least all the ones he can get via the Twitter API thanks to e.g. rtweet, in order to be able to look through them next time he needs an expert. And in the random sample of tweets he’s read, let’s hope he was exposed to a great diversity of science topics (and of scientists), although, hey, the health and life related stuff is the most interesting of course. Just kidding. I liked reading tweets about various scientists, science rocks! And these last words would be labelled with “performing arts”, perfect way to end this post.